
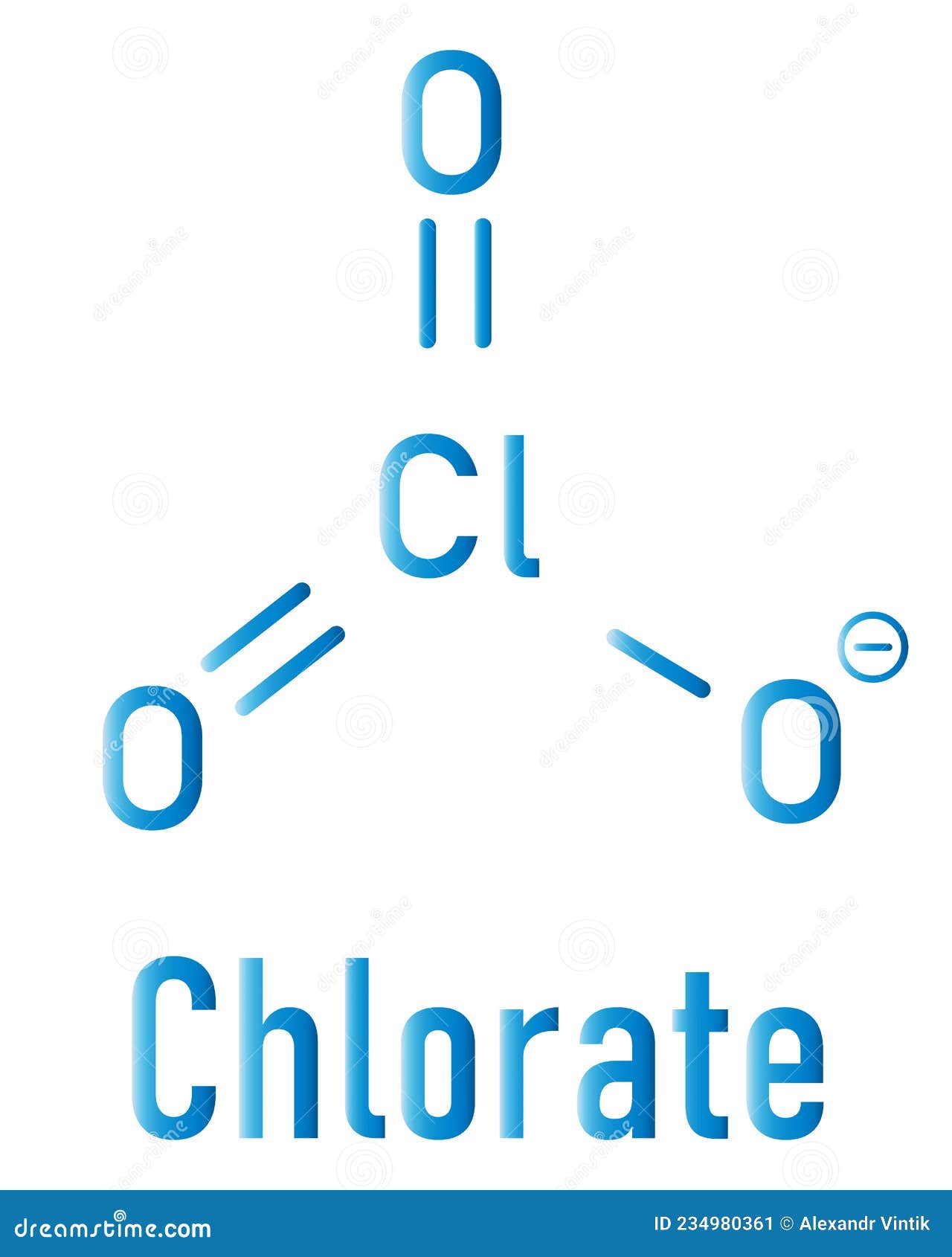
Marvin JS is ready for RESTful web services. Our sketcher can be exposed on any web browser supporting JavaScript and HTML5 technology. The public API provided with the editor enables web developers to seamlessly embed this application with their homegrown custom solutions (e.g.: adding custom buttons, custom templates, UI presets). Marvin JS has been designed for easy integration with web-based applications. Read more about Marvin JS features in the User Guide Easy to integrate and customizable – An exciting new interface You can rotate or mirror, whole or partial molecules as well as, represent your “creation” in 3D using the most appropriate display mode. Furthermore, the application offers a simple and clean interface for visualizing structural characteristics of previously drawn or imported molecules. New tools are introduced for complex drawing, so you are able to create more complex structures without continuously toggling between tools (e.g.: drawing tool, smart R-group tool, reaction tool). Marvin JS provides a great solution for drawing and modifying standard organic chemical and organometallic structures, reactions, electron transfer mechanisms, Markush-structures, and query molecules targeting different use cases. Marvin JS was created not only to meet requests of chemistry professionals, but to deliver solutions at the highest standard. 2D/3D clean, automap, stereo calculation). The Graphical User Interface and calculations are flexible: you can customize the application for your requirements, and extend its functionalities by installing the appropriate web services (e.g. It's seamlessly integrated into third-party web-based applications, and runs smoothly on all major browsers. Marvin JS provides quick and convenient ways to draw and modify standard and advanced chemical structures. sy2), Tripos Sybyl Line Notation (.sln), Beilstein ROSDAL (.ros), XYZ Files (.Designing molecules on the web - fast, smart and intuitive mmod), Schrödinger Maestro (.mae), Standard Molecular Data (.smd), Tripos Mol2 (.mol2. ent), RCSB Protein Data Bank Markup Language (.xml. mmcif), RCSB MacroMolecular Transmission Format (.mmtf), RCSB Protein Data Bank Files (.pdb. rd), MDL RXNFiles, both V2000 and V3000 connection tables (.rxn), MMI SketchEl Molecule (.el), Molinspiration JME String (.jme), RCSB Binar圜IF (.bcif), RCSB Macromolecular Crystallographic Information File (.cif. dx), ISIS Sketch File (.skc), ISIS Sketch Transportable Graphics File (.tgf), MDL MOLFiles, both V2000 and V3000 connection tables (.mol. smiles), IUPAC InChI (.inchi), IUPAC JCAMP-DX (.jdx. Read and write many popular chemical file types for working with the applications you use:ĪCD/ChemSketch Documents (.sk2), ChemDoodle Documents (.icl), ChemDoodle 3D Scenes (.ic3), ChemDoodle Javascript Data (.cwc.js), CambridgeSoft ChemDraw Exchange (.cdx), CambridgeSoft ChemDraw XML (.cdxml), Crystallographic Information Format (.cif), CHARMM CARD File (.crd), ChemAxon Marvin Document (.mrv), Chemical Markup Language (.cml), Daylight SMILES (.smi. Algorithmic Analysis of Cahn−Ingold−Prelog Rules of Stereochemistry: Proposals for Revised Rules and a Guide for Machine Implementation. and is 100% accurate in all 300 test cases provided. The CIP algorithm in ChemDoodle is validated against the test suite provided by Hanson et. Stereochemical features in your structures will be assigned "R", "S", "E", "Z", "M" and "P" descriptors. to remove any ambiguities and describe a completely consistent system for CIP assignments.ĬhemDoodle implements all 6 current CIP rules as well as auxilliary desciptors and mancude ring support. The most recent CIP rules from IUPAC were then algorithmically analyzed and standarized by Hanson et al. These rules were adopted by IUPAC for naming standards and fully described in the Blue books. While flawed, they have seen many revisions over the decades and were clarified by the work of Paulina Mata. The CIP rules have long been the standard for describing configurations of stereochemical features in a molecule.


 0 kommentar(er)
0 kommentar(er)
